Motivation
Within the community of cognitive scientists, the analysis of movement data is becoming more and more popular. While continuous measurements promise insights that reaction times cannot provide, researchers conducting their first tracking experiment may feel overwhelmed by the novel data handling requirements. Consequently, they often resort to out-of-the-box software restricting their analysis choices or poorly documented code snippets from colleagues. Thus, time is spend learning peculiarities of certain software (or colleagues) that could also be invested in learning overarching principles of data analysis or the development of own analysis routines.
The aim of mousetRajectory is to provide scientists with
an easy-to-understand and modular introduction to the analysis of
mouse-tracking and other 2D movement data. While
mousetRajectory should provide most functions needed to
analyze your first experiment, we strongly encourage you to extend
and/or replace certain modules with own code; a deeper understanding of
the analysis process will naturally lead to better interpretations of
the results. Therefore, we tried to make the source code as easy to
understand as possible even when this leads to slower function
execution. We further recommend to inspect and understand the functions
you are executing (if you are using RStudio on a Windows Machine, hit
F2 to inspect source code of functions).
Installation
You can install mousetRajectory from CRAN with
install.packages("mousetRajectory")Alternatively, you can keep up to date and install the latest development version of mousetRajectory from github.com/mc-schaaf/mousetRajectory with:
if(!require("devtools")){install.packages("devtools")}
devtools::install_github("mc-schaaf/mousetRajectory")Function Overview
Currently, the following functions are featured:
- Preprocessing:
-
is_monotonic()checks whether your timestamps make sense and warns you if they don’t. -
is_monotonic_along_ideal()checks whether your trajectories make sense and warns you if they don’t. -
time_circle_left()tells you the time at which the starting area was left. -
time_circle_entered()tells you the time at which the end area was entered. -
point_crosses()tells you how often a certain value on the x or y axis is crossed. -
direction_changes()tells you how often the direction along the x or y axis changes.
-
interp1()directs you to the interpolation function from the awesomesignalpackage. Thus, you do not have to calllibrary("signal"). Such time-saving, much wow. Also, not having to attach thesignalpackage avoids ambiguity betweensignal::filter()anddplyr::filter()in your search path. -
interp2()is a convenience wrapper tointerp1()that rescales the time for you.
-
- Spatial measures:
-
starting_angle()computes (not only starting) angles. -
auc()computes the (signed) Area Under the Curve (AUC). -
max_ad()computes the (signed) Maximum Absolute Deviation (MAD). -
curvature()computes the curvature. -
index_max_velocity()computes the time to peak velocity, assuming equidistant times between data points. -
index_max_acceleration()computes the time to peak acceleration, assuming equidistant times between data points.
-
- Other measures
-
sampen()computes the sample entropy.
-
A Simple Example
Let us assume we are conducting a simple experiment. In our setup, participants must respond by moving a mouse cursor from a starting position (bottom circle, coordinates (0,0)) to one of two end positions (top circles, coordinates (-1,1) and (1,1)):

dat stores simple toy data that may reflect our pilot
results. Let’s inspect the structure of the data:
head(dat)
#> # A tibble: 6 × 5
#> Trial Target Time x_coord y_coord
#> <int> <chr> <int> <dbl> <dbl>
#> 1 1 left 0 0 0
#> 2 1 left 1 -0.01 0.0140
#> 3 1 left 2 -0.02 0.0278
#> 4 1 left 3 -0.03 0.0416
#> 5 1 left 4 -0.04 0.0554
#> 6 1 left 5 -0.05 0.069
# gg_background has been created previously and is a ggplot object
gg_background + geom_path(aes(x_coord, y_coord, group = Trial), dat)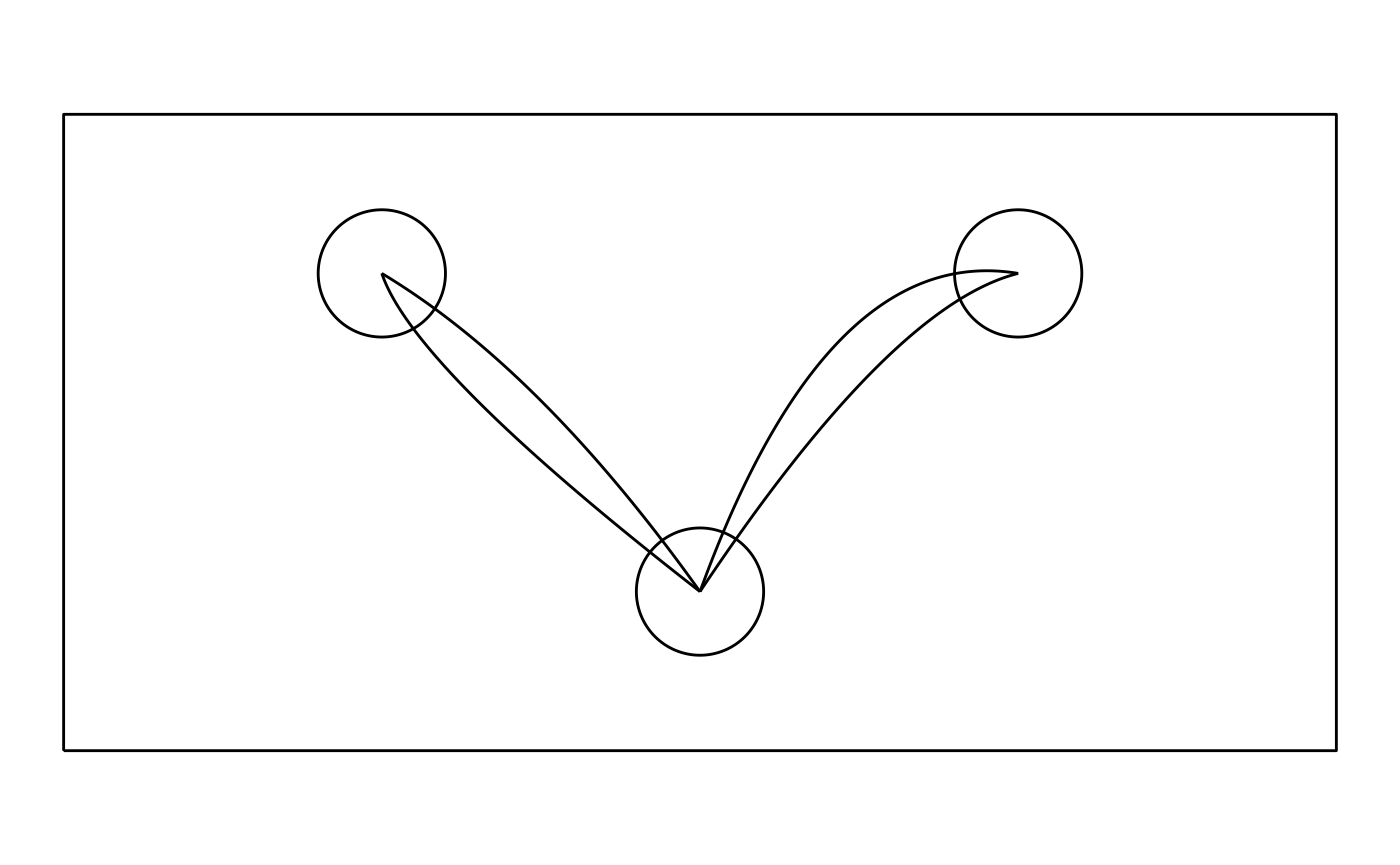
In this example, Time reflects the time passed since the
onset of the imperative stimulus and always increases by 1 arbitrary
unit. In a real experiment, a sensible first pre-processing step would
be to double-check the order of your data via
is_monotonic() applied to Time. Further, in a
real experiment you would likely group by not only by Time,
but also other variables like a subject or block identifier.
dat <- dat %>%
group_by(Trial) %>%
mutate(
# will throw a warning if times are not monotonically increasing
is_ok = is_monotonic(Time)
)As a next step, we will recode the coordinates so we can treat
movements to the left and to the right in the same way. This enables us
to restrict the trajectories to their relevant parts, i.e., after the
home was left and before the target has been reached. This also allows
us to extract our first dependent measures, InitiationTime
and MovementTime:
dat <- dat %>%
group_by(Trial) %>%
mutate(
x_coord = ifelse(Target == "left", -x_coord, x_coord),
InitiationTime = time_circle_left(
x_coord,
y_coord,
Time,
x_mid = 0,
y_mid = 0,
radius = 0.2
),
CompletionTime = time_circle_entered(
x_coord,
y_coord,
Time,
x_mid = 1,
y_mid = 1,
radius = 0.2
),
MovementTime = CompletionTime - InitiationTime
) %>%
filter(Time >= InitiationTime & Time < CompletionTime)
dat %>%
group_by(Trial, InitiationTime, CompletionTime, MovementTime) %>%
count()
#> # A tibble: 4 × 5
#> # Groups: Trial, InitiationTime, CompletionTime, MovementTime [4]
#> Trial InitiationTime CompletionTime MovementTime n
#> <int> <int> <int> <int> <int>
#> 1 1 12 84 72 72
#> 2 2 10 76 66 66
#> 3 3 10 79 69 69
#> 4 4 9 83 74 74
gg_background + geom_path(aes(x_coord, y_coord, group = Trial), dat)
Note that filtering the data to the relevant part leads to an unequal
amount of data points for each trajectory (column n). This
is bad news when you want to display average trajectories! One solution
for this problem is “time-normalization,” i.e., a separate linear
interpolation of the x and y coordinates at certain time points. It has
been proposed that this process should be done prior to the computation
of MAD, AUC, etc. So let’s do this via interp2(), a
convenient wrapper to signal::interp1():
dat_int <- dat %>%
group_by(Trial) %>%
reframe(
Time_new = 0:100,
x_new = interp2(Time, x_coord, 101),
y_new = interp2(Time, y_coord, 101),
)Now we are ready to compute dependent measures like AUC and MAD:
dat_int %>%
group_by(Trial) %>%
summarise(
MAD = max_ad(x_new, y_new),
AUC = auc(x_new, y_new),
CUR = curvature(x_new, y_new)
)
#> # A tibble: 4 × 4
#> Trial MAD AUC CUR
#> <int> <dbl> <dbl> <dbl>
#> 1 1 0.0353 0.0239 1.00
#> 2 2 -0.0274 -0.0183 1.00
#> 3 3 0.0462 0.0314 1.01
#> 4 4 0.126 0.0907 1.04As a last step, you may want to plot your average trajectory:
dat_avg <- dat_int %>%
group_by(Time_new) %>%
summarise(
x_avg = mean(x_new),
y_avg = mean(y_new)
)
gg_background +
geom_path(aes(x_avg, y_avg), dat_avg) +
geom_path(aes(c(0, 1), c(0, 1)), linetype = "dashed") # ideal trajectory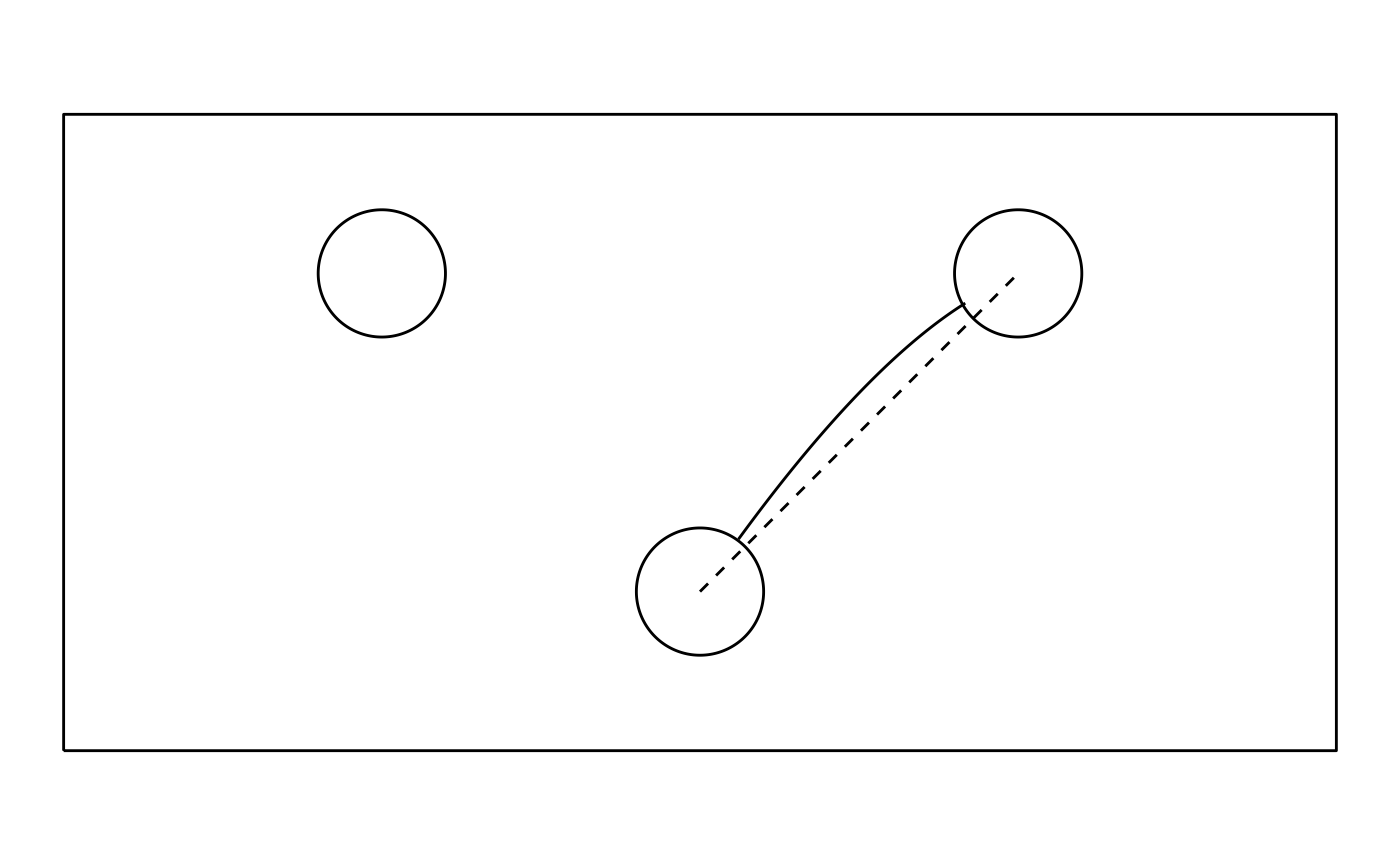
Congratulations, you worked trough your first mouse-tracking analysis!
As a final note: This package is currently under development. If you spot any bugs or have other improvement suggestions, please let us know by filing an issue at github.com/mc-schaaf/mousetRajectory/issues.